Monday Links: Model Interpretation, long range enhancers, designing DNA and more
Some interesting reads to start your week
Apologies for the slow posting this fall. Several pieces on fascinating topics are in the pipeline but various obligations have kept me from finishing them. Stay tuned.
In the meantime, here are six links that are worth reading at the beginning of your week.
Breaking into the black box of deep learning
Deep learning is most often used for making predictions or classifications (is there a cat in this image?), and for generating content, like the answers to coding questions that ChatGPT generates for my stats and coding class. Deep learning models are good at this because they are highly proficient at learning patterns in high dimensional data. And folks are increasingly interested in finding out just what patterns were learned, rather than merely being satisfied with accurate predictions. The literature on interpreting machine learning models is moving quickly. Neel Nanda put together a great reading list of important papers on this topic. Check it out for important work on transformers, attention, superposition, sparse autoencoders, etc.
Visualizing Transformers
Speaking of black boxes, what exactly is a transformer? If you’re new to this subject, 3Blue1Brown has a new set of excellent video lessons that tour the inside of LLMs. If you’re not familiar with 3Blue1Brown, you are missing out. The site features high quality introductory videos on math, physics, and machine learning. If you need a quick primer on a Fourier series or backpropagation, this site is for you. (For these first cool two links, hat tip to Brad DeLong’s Grasping Reality substack. I liked them so much that I needed to share.)
A genomic range extender
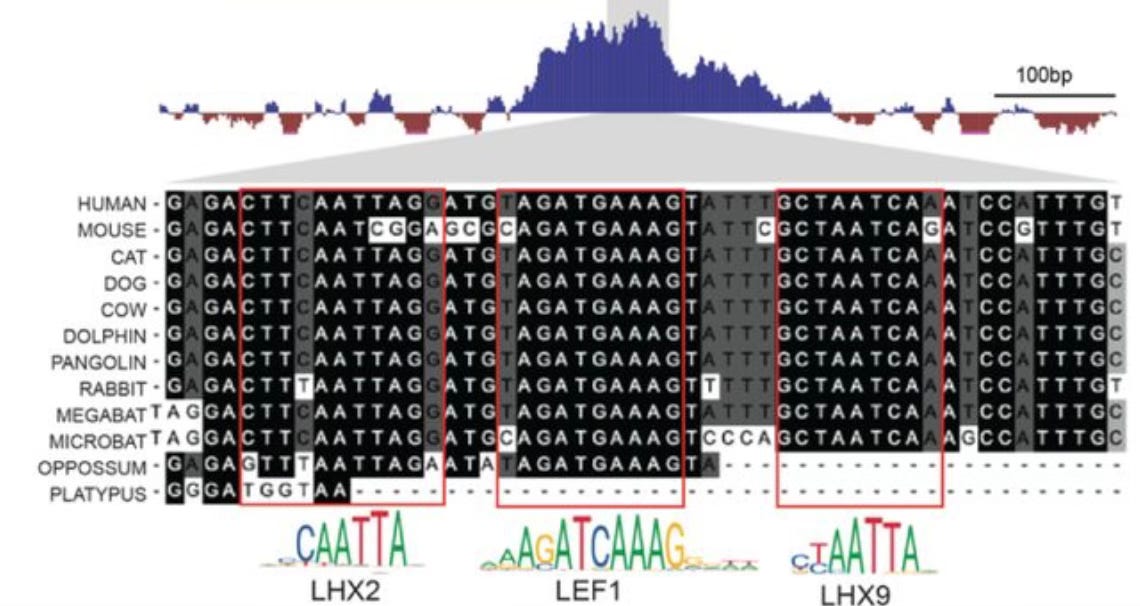
Unlike the wi-fi range extender in my house, which is quickly becoming obsolete, a genomic range extender sequence discovered by Evgeny Kwon and colleagues is astonishingly well-conserved. How regulatory DNA sequences like enhancers act over long genomic distances is a questions that has received a great deal of interest, but the features of a sequence that confer long-distance activity are still generally mysterious. Kwon an colleagues found a short sequence, composed of three protein binding sites, that can increase the genomic range of an enhancer by almost ten fold. The sequence itself does not act as an enhancer, but it is, remarkably for a regulatory sequence, conserved across nearly 180 million years of evolution — it is present in marsupials and placental mammals.
Designing DNA: Which model?
On the theme of regulatory DNA and machine learning, we are seeing the a growing wave of generative DNA models. I will cover this topic in detail in a future post, but a preprint by Cold Spring Harbor computational biologist Peter Koo has a nice mini-review of the field in its introductory section and an appendix. In the paper, Koo and his lab introduce a diffusion modeling approach for designing DNA. Read Appendix A for a brief discussion of generative adversarial networks, variational autoencoders, normalizing flows, gradient ascent, simulated annealing, etc., in DNA design. If you want to get up to speed on the literature on generative models of DNA, this is a great place to start.
“Chaperoneopathies”: Brain disorders and pathogenic variants in chaperone complexes
A paper in the Nov. 1 issue of Science reports pathogenic variants in seven of the eight subunits of the TRiC protein-folding complex. These pathogenic variants are all associated with brain malformations, intellectual disability, and seizures. This is yet another example of the mysterious phenomenon of an organ-specific disease caused by variants in general purpose proteins. Variants in different TRiC subunits affect the complex differently, and lead to different disease phenotypes. Part of this work was done by my WashU colleagues at the Children’s Discovery Institute, which has a great team focused on detailed functional characterization of candidate variants.
30 years of genome technology development
Bob Fulton has seen it all. Our MGI director of technology development has worked on genome technologies since the early days of the human genome project, and he took a few minutes to share his thoughts on the changes he’s seen over the years.